|
Supervised by: Courtney Hanna (cwh36@cam.ac.uk) & Miguel Constancia (jmasmc2@cam.ac.uk) |
|
|
Project Title |
Investigating the role for epigenetic modifier MLL2 in placental development |
|
Project description The placenta forms the maternal-foetal interface in pregnancy, controlling nutrient and waste exchange, hormone production and immunotolerance between mother and foetus. Impaired placental function has been widely linked to poor pregnancy outcomes. However, we currently have little understanding of the early regulators of placental development that lead to a functional placenta and healthy pregnancy. After implantation, embryonic stem cells undergo a process of ‘epigenetic priming,’ a necessary transition that allows these pluripotent cells to be receptive to signalling for differentiation into all of the cell types of the body. A hallmark of this priming event is the deposition of bivalent chromatin at a subset of promoters, marked by active H3K4me3 and repressive H3K27me3, poising them for activation (Bernstein et al. 2006). Failure to prime the embryonic genome results in developmental defects. The role for bivalent chromatin in placenta remains contentious (Rugg-Gunn et al. 2010; Andergassen et al. 2019; He et al. 2020) and hence is an important research question. The aim of this project is to characterise and test the functional importance of epigenetic priming in placental cells, linking poor priming to defects in placentation and consequences on foetal growth. Using mouse as a model, this project will investigate the role for H3K4me3 methyltransferase MLL2, one of the key enzymes responsible for establishing bivalent chromatin (Denissov et al. 2014), in priming placental cells for differentiation. Three important questions will be addressed:
The student will obtain training in a wide variety of experimental techniques including next generation sequencing, histology, embryology and bioinformatics.
|
|
|
References
|
|
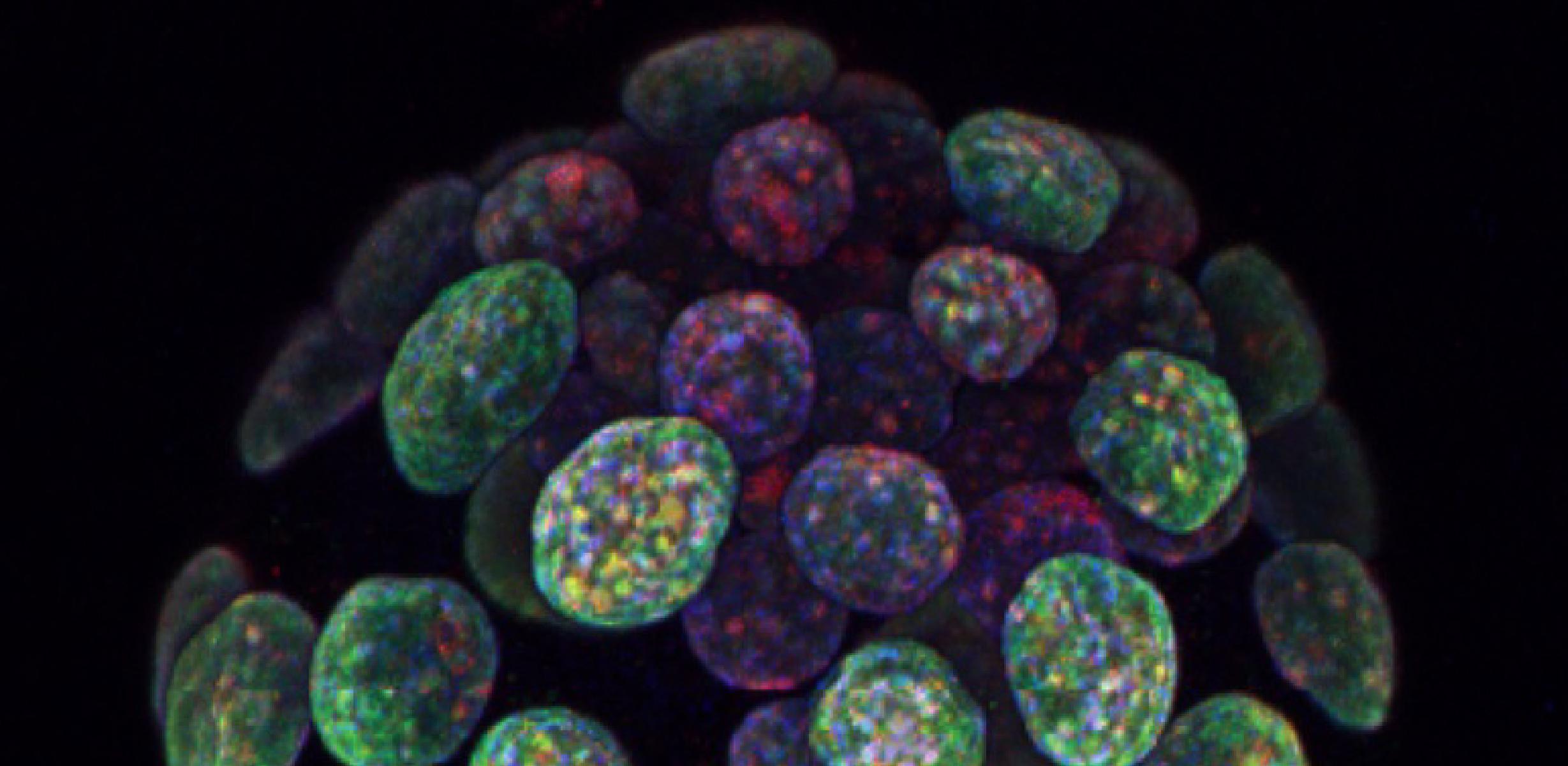
Image caption: A pre-implantation blastocyst-stage mouse embryo contains two cell types, the outer trophectoderm layer (which later forms the placenta) and the inner cell mass (which later forms the foetus). During epigenetic reprogramming in early embryogenesis, the cells from the trophectoderm and inner cell mass acquire different patterns of epigenetic modifications, such as DNA methylation (red) and histone 3 lysine 9 dimethylation (green). The programming of epigenetic modifications is essential for gene regulation and lineage specification during development. Image provided by Fátima Santos (Epigenetics Programme, Babraham Institute)

