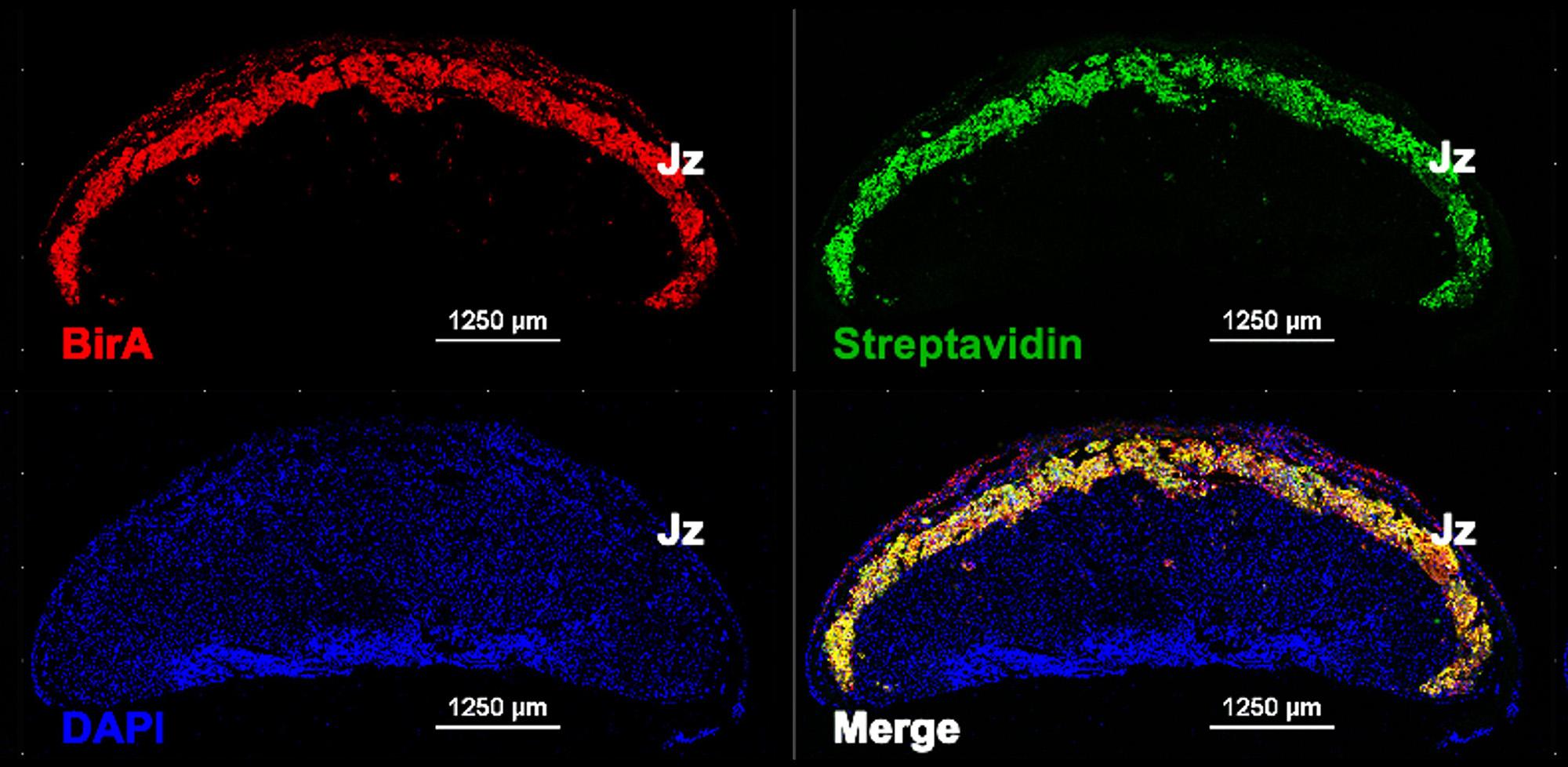
Image: In vivo labelling of placental endocrine cells with biotin in BirA*G3 mice as a novel tool to follow trafficking of secreted proteins to maternal and fetal organs. Immunofluorescent staining of E18.5 mouse placental sections with: (1) Anti-BirA (red) – demonstrating expression of transgenic biotin ligase BirA in the placental junctional zone – Jz), (2) -Streptavidin (green) – demonstrating endogenous biotinylation of proteins in Jz by the activity of BirA, and (3) DAPI (4′,6-diamidino-2-phenylindole - blue) – a fluorescent stain that binds to DNA. Image credit: Ionel Sandovici, Miguel Constancia lab.
|
Supervised by: Prof. Miguel Constancia (jmasmc2@medschl.cam.ac.uk) and Prof. Sue Ozanne (seo10@cam.ac.uk) |
|
|
Project Title: Metabolic determinants of early placental development |
|
| Host Department: Department of Obstetrics & Gynaecology and Institute of Metabolic Science | |
|
Project description Maternal physiology undergoes extensive changes during pregnancy to ensure an adequate supply of nutrients to the fetus. One of these adaptations, occurring in late pregnancy, is increased insulin resistance (i.e. decreased sensitivity of maternal organs to insulin-mediated glucose uptake). The development of maternal insulin resistance reduces glucose uptake and energy storage in maternal skeletal muscle, increases hepatic gluconeogenesis and increases lipolysis in the adipose tissue. This promotes glucose, amino acids and lipid transfer to the growing fetus. In response to maternal insulin resistance, the capacity of maternal pancreatic cells to produce insulin is increased (e.g. through beta-cell mass expansion). It is thought that maternal peripheral insulin resistance is largely orchestrated by the placenta, a specialized organ at the interface between the mother and the fetus, and the site for maternal-fetal exchange. It is well established that the placenta regulates maternal physiological adaptations, including development of insulin resistance, through the secretion of hormones and signalling factors into the maternal circulation. However, the complete profile of these placental-derived secreted factors, the precise organ they target and the mechanisms by which they modify organ physiology, e.g. insulin sensitivity, are largely unknown. The student will use newly developed mouse genetic tools to elucidate placental-to-maternal communication during normal and complicated pregnancies. Specifically, this project has 3 aims:
This project is likely to identify novel biomarkers of insulin sensitivity/resistance in pregnancy, which is highly relevant to pregnancy complications such as gestational diabetes, that affects 1 in 7 pregnancies globally. In addition, it will define the role of the placental factors in the control of glucose homeostasis during pregnancy, with potential therapeutic applications. |
|
|
References
|
|

