|
Supervised by: Prof Steve Charnock-Jones (dscj1@cam.ac.uk) and Dr Irving Aye (ia319@medschl.cam.ac.uk) |
|
|
Project Title: Metabolic determinants of early placental development |
|
| Host Department: Department of Obstetrics & Gynaecology | |
|
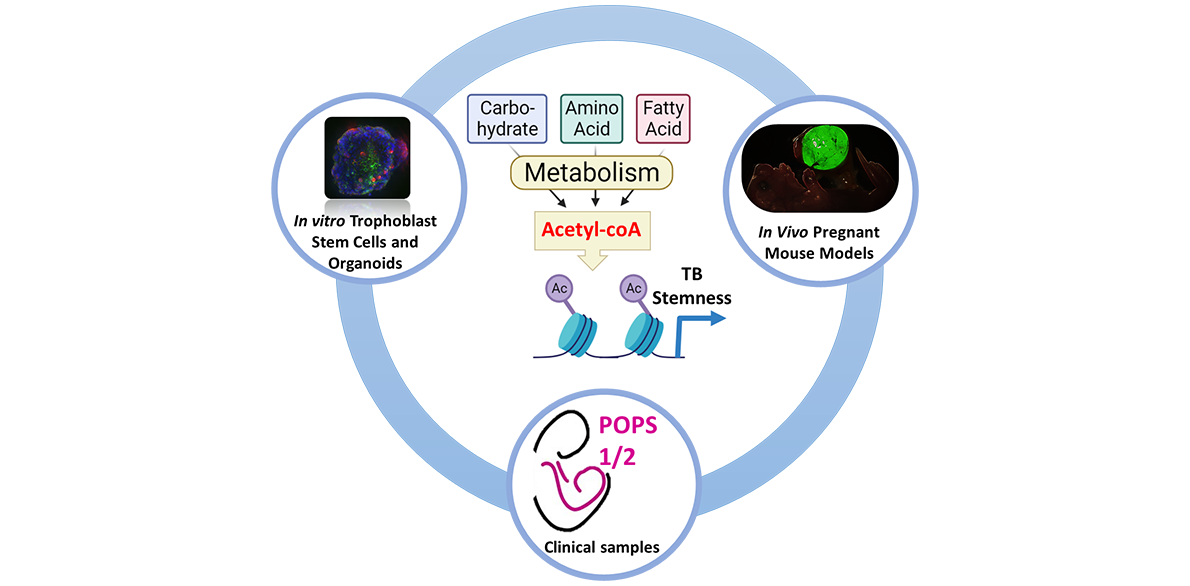
Project description During early development, placental trophoblasts face diverse metabolic challenges including nutrient and oxygen-limited microenvironments and changing environmental conditions. It is also during this stage that vast changes in gene expression coincide with reprogramming of cellular metabolism. However, it is unclear how trophoblasts assess and respond to their metabolic resources to achieve a coordinated effect on cell proliferation, differentiation, and placental development. Cell metabolism is conventionally viewed as a means of obtaining bioenergy for cellular homeostasis or building blocks for biomass growth. However, metabolic intermediates can also strongly influence gene expression through additional non-bioenergetic functions, for instance as rate-limiting substrates or co-factors in a variety of epigenetic processes. This project aims to understand the mechanistic links between cell metabolism and epigenetic programs directing trophoblast stemness and differentiation. Key questions to be addressed are: How does the placenta respond and adapt to changes in varying environmental conditions? How does cellular metabolism influence epigenetic networks in placental development? The underlying hypothesis is that trophoblast metabolism directs gene expression programs regulating stemness or differentiation by controlling the availability of metabolic intermediates necessary for epigenetic processes. Leveraging on the expertise of both the supervisors’ labs, the candidate will receive training in trophoblast biology, metabolism and computational biology. The candidate will utilise human trophoblast stem cells and organoids, as well as primary trophoblasts from term placentas. This work may be complemented by mouse models of placenta-specific gene manipulation. Additionally, training will be provided in the following state-of-the-art research techniques:
This project will explore the unchartered field of “Placental Metabo-Devo”, i.e. the different mechanisms by which metabolism impacts on the processes governing placental development. Addressing these fundamental biological questions may help us understand novel mechanisms of placental-related pregnancy complications such as miscarriage, preeclampsia and fetal growth restriction. |
|
|
References
|
|

